Anything launched to space is meticulously tested, monitored, and often sanitized. However, our control over spacecraft and the items aboard is not total, especially at the microscopic level. Microbial hitchhikers still have the capability to catch a ride to space, with either the cargo or the crew.
While people literally could not live without these tiny organisms, many of which are benign or beneficial, there is still a possibility that the assortment of microbial hitchhikers could include disease-causing pathogens that might make the crew sick. That is why scientists use the International Space Station as a testing ground to study how to keep astronauts safe and healthy on long-duration missions. These studies also benefit humans on Earth by providing a better understanding of how microbes behave in a sanitized, isolated, and confined environment.
This work is also forming a new area of scientific research. Many microbes can be stressed by the conditions of space, and some respond with genes encoding resistance to antimicrobial agents. It is therefore important to understand what types of space-residing microbes are present in the environment and whether they can become impervious to sanitation techniques.
“The goal is to enable near-real-time microbial diagnostics of the space station environment,” said Sarah Wallace, a microbiologist in the Biomedical Research and Environmental Sciences division at NASA’s Johnson Space Center in Houston. “We are looking at what is in the air, on surfaces, and in the water that the crew is surrounded by all day, every day.”
Known, worrisome bacteria can be identified by their unique biological blueprint, contained within molecules of deoxyribonucleic acid (DNA). DNA is made up of four base molecules that link together to encode instructions for cell growth and behavior. Identifying the order of the bases using the process of DNA sequencing clues researchers in to the identity of the organisms and how they might behave.
As the studies continue taking measurements, the data can expose patterns that exist in microbial composition on the space station. “It is kind of like taking a census,” said David J. Smith, acting chief of the Space Biosciences Division at NASA’s Ames Research Center in California’s Silicon Valley. “The demographics data collected can be used years down the road for planning decisions about how we coexist with microbes in spacecraft.”
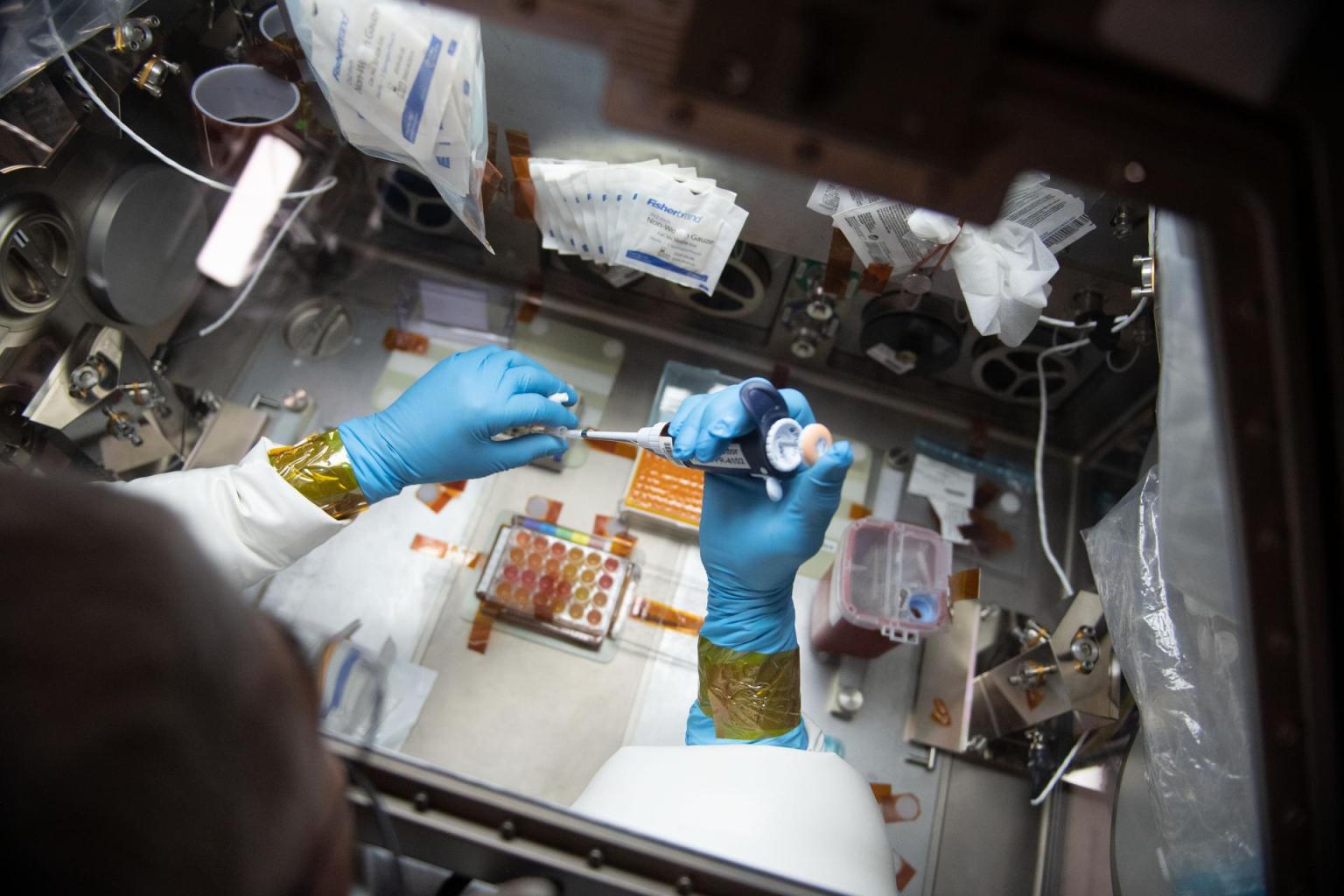
However, taking the census can be complex. The equipment required for DNA sequencing has historically been expensive and time intensive and has required specialized expertise to operate, limiting its use in space. For example, a series of NASA microbial tracking experiments and the 3D Microbial Monitoring study had to rely on astronauts collecting hundreds of samples by wiping down selected surfaces with swabs, packing the samples in plastic bags, and sending them back to Earth for identification with DNA sequencing facilities.

Sending samples from the orbiting lab back to Earth is complicated and adds significant time between the initial sampling and identifying the microbes present. The quicker we can identify unknown microbes on the International Space Station, the more prepared the crew will be to respond.
The key is finding a way to do complex microbial ecology surveys in space.
Microgravity and the restrictions on size and chemicals that can be safely used in the enclosed environment of the space station add to the challenge of designing a DNA experiment for space.
“We broke it apart at every angle and looked at how we can use the least amount of and safest tools and substances and get the best science,” said Wallace. “That is where we go outside of the box and come up with a different solution.”
Thanks to the work of Wallace and her team, sample return could soon be a thing of the past for microbiology studies.
In 2016, NASA astronaut Kate Rubins successfully conducted the first DNA sequencing in space, while researchers on the ground performed the same experiment, opening the door to molecular biology research in spaceflight conditions. The team used a MinION (pronounced “min ion”) sequencing platform, a device no bigger than a cell phone, to read the nucleic acid bases in samples sent to the station for study. This technology can enable scientists to quickly identify pathogens on the space station or on future exploration missions, and even potentially identify life on other planets in the solar system if it shares a common biochemistry with life as we know it on Earth.

“Our first sequencing demonstration in space was a big deal because the work was done from concept to flight in less than a year. Traditionally, it takes a much longer time to get a new instrument on station,” said Smith. “We showed it is possible to be nimble in our science as we go beyond low-Earth orbit with future experiments.”
The initial work of DNA sequencing opened the door to using other molecular biology methods in space. “Having an entire molecular laboratory in space is just going to explode what we can do there,” said Wallace.
And explode it has. Astronauts used a genome editing technique called CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) for the first time in space significantly expanding the space station’s molecular biology toolkit once again by enabling studies of DNA repair. Numerous students have sent DNA research to space through the Genes in Space program. Scientists have even used MinION to fully identify unknown microbes on the orbiting laboratory without returning samples to Earth. The ability to identify microbes in space could aid in the diagnosis and treatment of astronaut ailments in real time. NASA astronaut Ricky Arnold even deployed the Biomolecule Sequencer with a swab method that did not require growing the bacteria prior to sequencing. With this direct “swab to sequencer” method, the process becomes faster and simpler for crew members to identify microbes onboard the station. This method is now being evaluated to replace culture-based microbial monitoring that is currently conducted aboard the space station.
While optimized microbiology procedures will help enable deep space exploration, developing an autonomous molecular laboratory is expected to benefit humanity on Earth as well. This could be especially true for remote communities and hospitals without a lot of technology or funding.
“The ability to generate near-real-time molecular microbial data could provide a huge benefit toward understanding and responding to the antimicrobial resistance crisis and in resource-limited environments,” said Wallace. “It is great to see the parallels between our work and the work by other researchers that are implementing these tools in other diverse places. If you can do these types of investigations on the ocean floor, on the space station, or on a table in a remote village, you can do them anywhere.”
Designing simple procedures that can be performed by crew members who may not be microbiologists and by workers anywhere who may not have genetics expertise broadens the capacity for disease testing in underdeveloped countries or remote settings on Earth.
The NASA research team has gone on to collaborate with the Centers for Disease Control, looking into how streamlined procedures developed for the space station can be implemented to determine the cleanliness of hospital rooms. Using these microbial tracking methods in hospitals could provide a much faster turnaround time, even when implemented by untrained individuals. This capability might give hospitals the tools to detect microbes that may be resistant to antibiotics that could be present even after cleaning.
“Surprises are embedded in basic research and exploration,” said Smith. “It could be establishing a more efficient workflow for microbial identification or a useful pattern revealed in space that would otherwise be hidden on Earth. We never fully know what we are going to discover in spaceflight and the potential benefits to humanity – and that is why it is worth doing.”
Additional Resources:
Explore the evolution of DNA sequencing in space
Space Station Microbiology: Where People Go, Microbes Follow