Gravitational Effects and Radiobiology of Microbes in Space
(GERMS)
Microbial Eco-Evolutionary Dynamics in Microgravity
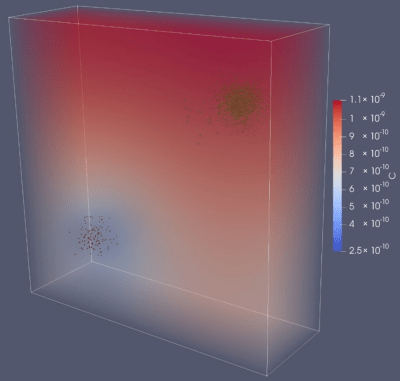
Altered gravity is a unique feature of the physical environment of space: reduced gravity on the surfaces of the Moon and Mars, and microgravity in orbit. Most microbes are too small to sense the force of gravity directly, but one way they may experience its effects is through its influence on their fluid environment. On Earth, gravity drives fluid mixing via density-driven convection; less gravity means less convective mixing. Everywhere microbes are found, they tend to live in multispecies communities, and commonly depend on chemical exchange through their environments to communicate and/or share resources. We are studying the potential effects of microgravity on multispecies communities using rotating wall vessels (RWVs) in the lab and are particularly interested in understanding the fidelity of microgravity simulation and the effects of experimental parameters such as rotation rate on that fidelity. We are also collaborating with the Computational Materials Group to build a simulation of the physical, biological, and chemical interactions of microorganisms in liquid culture in space, called CAMDLES: computational simulation of microbial populations in spaceflight (read about an early version of this model here.) This work has been funded by a grant from the NASA Space Biology Program and the Ames Center Innovation Fund.
Point of Contact: Jessica Audrey Lee
Deep-space radiation effects on microbes at the single-cell level
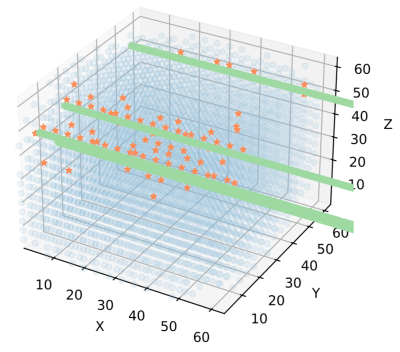
Space radiation comes in many different flavors. Charged-particle radiation in the form of Galactic Cosmic Rays (GCR) and Solar Particle Events (SPE) pose a substantial health risk to organisms traveling beyond Low Earth Orbit. However, for a tiny microbial cell, getting hit by a radiation particle can actually be a relatively rare event. What does this mean for our understanding of the effects of radiation on microbes, and how does that differ from what we know about radiation effects on multicellular organisms? Studying radiation from the perspective of individual microbial cells has implications not only for our ability to design microbial bioproduction systems to sustain human life in deep space, but also for our ability to interpret spaceflight radiobiology experiments using microbes as model organisms.
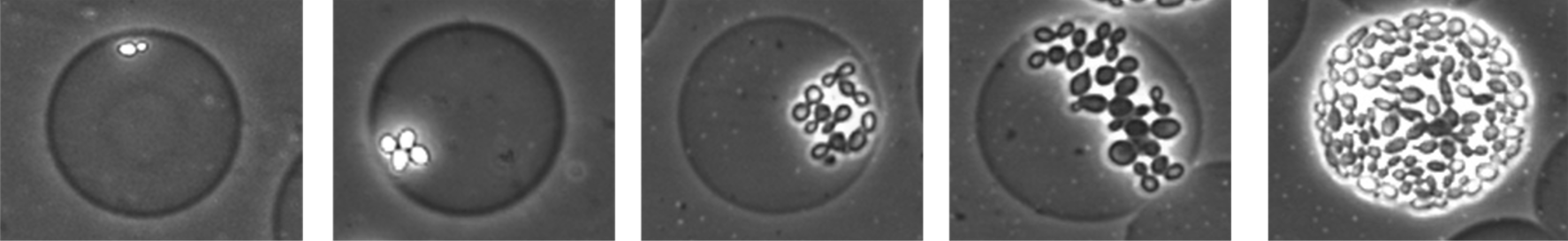
We are using diverse wet-lab methods to study single-cell radiation effects in yeast, including a collaboration with the Di Carlo Lab at UCLA to encapsulate yeast cells in hydrogel particles to facilitate lineage tracking. We have also developed a computational model to simulate yeast population growth in the presence of radiation, called AMMPER: Agent-based Model of Microbial Populations Exposed to Radiation (open-source and available at NASA GitHub). This work has been supported by MUREP, Space Grant, and the Ames Research Innovation Award.
Adaptive Evolution Under Simulated Microgravity
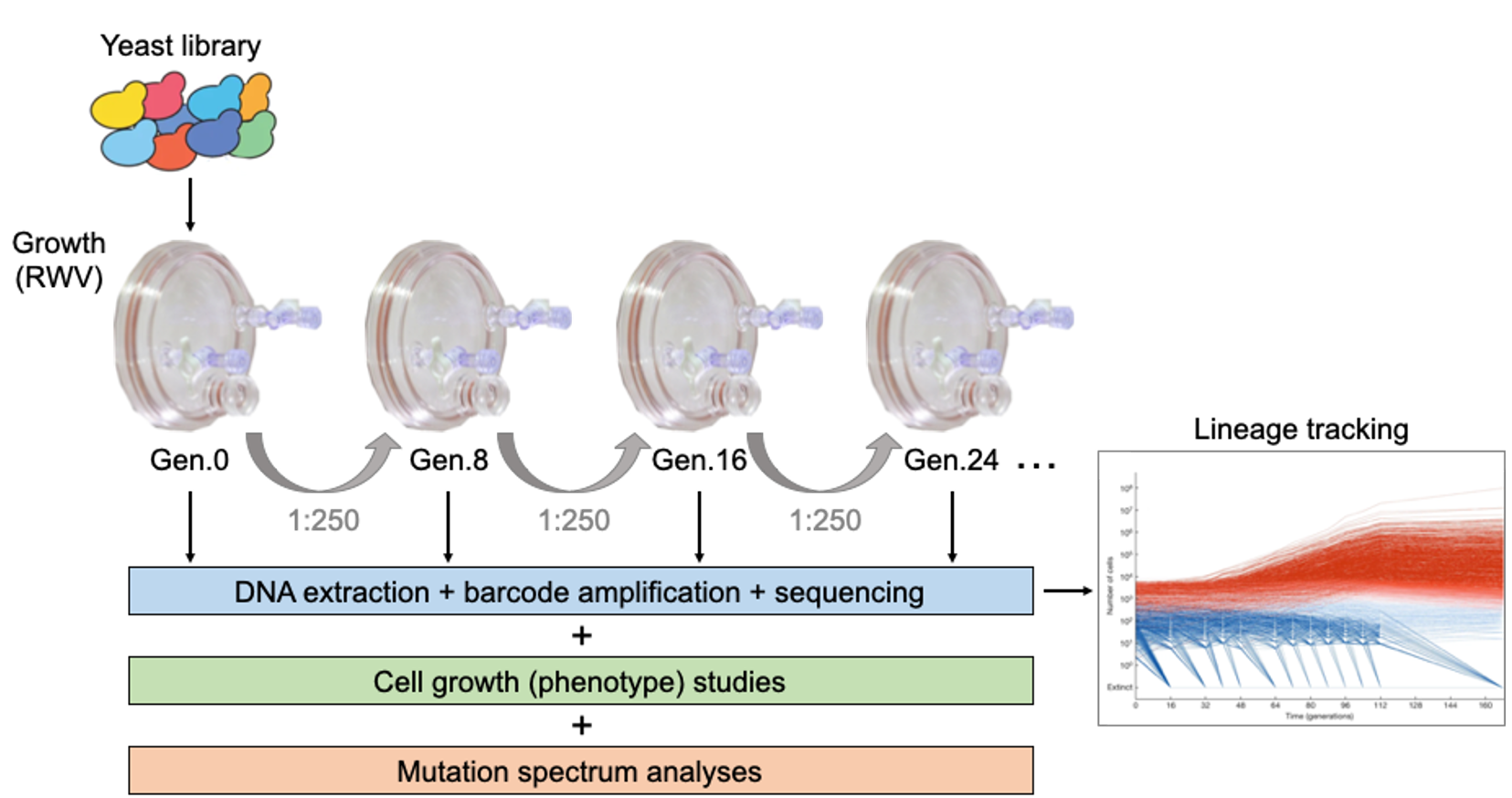
Adaptive evolution plays a major role in the appearance of new phenotypes in nature and is of key importance in biomedical research, including the onset of carcinogenesis and in the emergence of drug-resistant microorganisms. The harsh environment of space provides the ideal environment for the acquisition of adaptive mutations, particularly in microbial organisms. Experimental evolution studies in the laboratory allow us to better understand how specific mutations arise under spaceflight conditions. Our primary goal is to investigate how a eukaryotic organism responds to simulated microgravity after multiple generations. S. cerevisiae or budding yeast is a single-cell model eukaryote that has been widely used in biomedical research, primarily due to its well-characterized biology, easy manipulation, short division time, and, most importantly, its similarities with human cell functions. We hypothesize that beneficial mutations acquired through multiple generations under simulated microgravity will render cells with increased growth and/or metabolic rates, and phenotypes different to those observed in 1g controls. Studies like the one proposed here will allow us to identify beneficial mutations responsible for an adaptive response to space-like conditions.
This project is a collaboration with the Sherlock Lab at Stanford University. Dr. Sherlock’s team has developed barcoded yeast libraries, and the different DNA manipulation and next-generation sequencing approaches that are being used in this study. Once completed, all project data will be archived in the NASA GeneLab repository. This project has been funded by a grant from the NASA Space Biology program to Principal Investigator Sergio Santa Maria.